Sanghyuk Jung
Novel polygenic risk score approach with transcriptome-based weighting for genetic risk prediction of late-onset Alzheimer’s disease
Presenter

Visiting Ph.D. student
Kim Lab, Department of Biostatistics, Epidemiology & Informatics, The Perelman School of Medicine, University of Pennsylvania
Ph.D. candidate
Won Lab, Genomics & Digital Health Convergence Laboratory, The Samsung Advanced Institute for Health Sciences & Technology, Sungkyunkwan University
Abstract
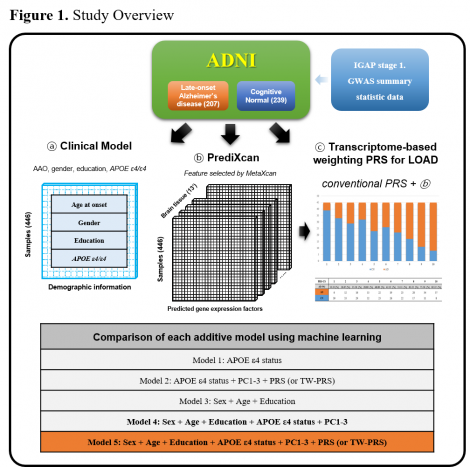
Background: Recent genetic studies showed that polygenic risk score (PRS) could be used to identify individuals at high risk of Alzheimer's disease (AD). Despite this success, prediction and early intervention of AD still remain challenging. In this study, we suggest a novel PRS approach that is weighted by predicted tissue-specific gene expression levels.
Method: We used whole-genome sequencing and phenotypic data of 446 European participants (207 late-onset AD cases and 239 cognitively normal controls) from the Alzheimer’s disease Neuroimaging Initiative (ADNI). We performed transcriptome-wide association studies (TWAS) in 13 brain regions by using MetaXcan (weights from GTEx V8) and GWAS summary statistics (IGAP stage 1 excluding ADNI participants) and integrated these 13 TWAS results using MultiXcan. To generate transcriptome-based weighting (TW)-PRS, expression weights in each gene were mapped to SNPs in the gene, and these were applied as additional weights to the SNPs’ beta coefficients in the GWAS summary statistics.
Result: An AD prediction using conventional PRS yielded a pseudo-R2 of 0.0462 (P=1E-04). Compared with the conventional PRS, the TW-PRS improved the performance and statistical power with a pseudo-R2 of 0.0574 (P=1.74E-05). A fully adjusted model achieved an AUC of 0.794 in which TW-PRS were significant even when APOE ε4 status and demographic information were adjusted (P=2.33E-05).
Conclusion: Our finding suggests that tissue-specific transcriptomic factors may be independent and complementary to conventional PRS and provide additional information for tissue-specific regulatory effects in AD.
Keywords
Transcriptome-based weighting polygenic risk score; Polygenic risk score; Transcriptome-wide association study; Alzheimer’s disease.Commenting is now closed.
About Us
To understand health and disease today, we need new thinking and novel science —the kind we create when multiple disciplines work together from the ground up. That is why this department has put forward a bold vision in population-health science: a single academic home for biostatistics, epidemiology and informatics.
© 2023 Trustees of the University of Pennsylvania. All rights reserved.. | Disclaimer


Comments